-
General Information -
How to Use: Example with Adrenaline -
How to Use: Example with PDB 3E6I -
Workflow -
References -
Problem In Running Application?
Example With 3E6I
Step 1) Enter PDB ID
Write down the PDB ID (Code) for Protein-ligand complex in the provided text field and hit the "Request data" button.
Step 2) Retrieved Ligands for 3E6I
Two ligands reported in PDB for CPY450 (3E6I) are shown below. Second ligand, indazole used for the similarity search.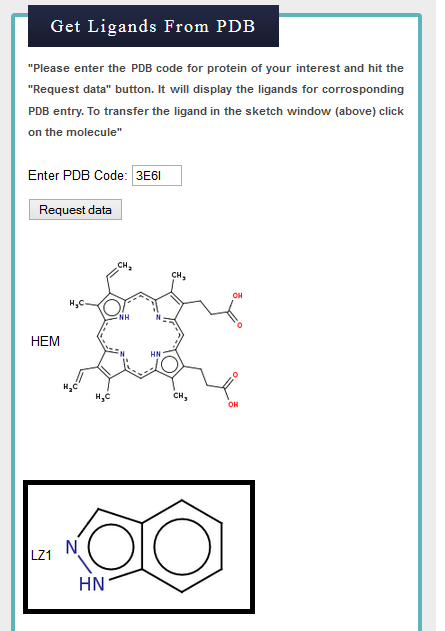
Step 3) Transfer indazole to msketch window
Click on the indazole molecule (number 2) to trasnfer structure to the msketch window.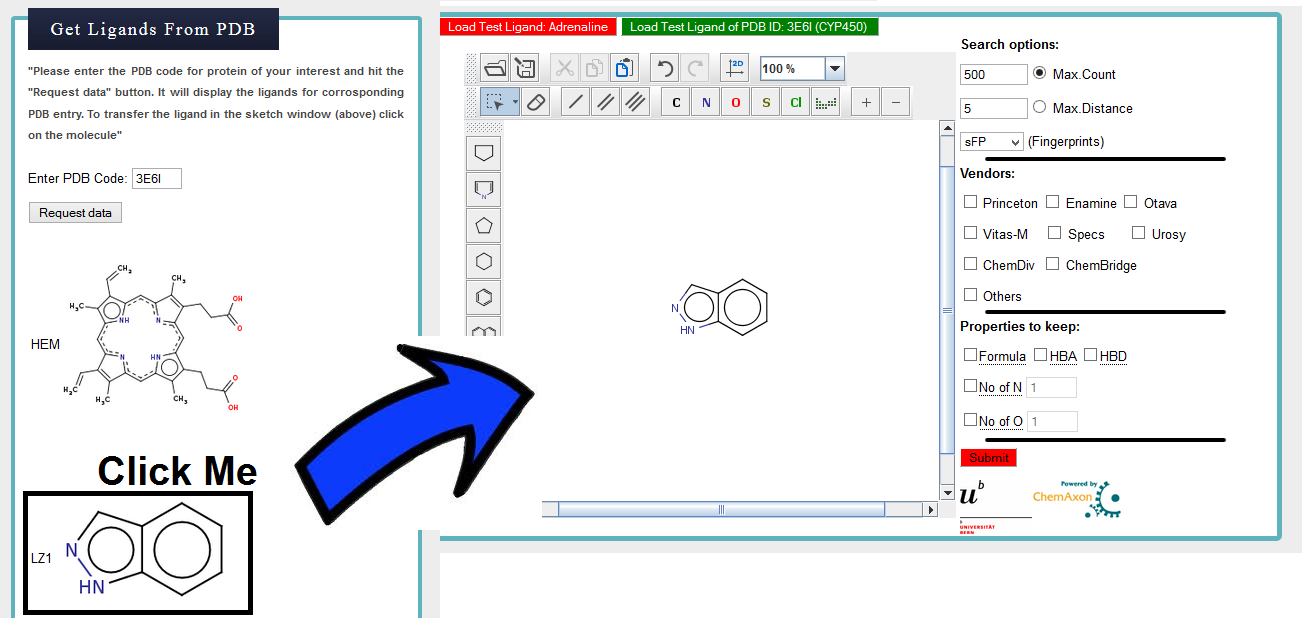
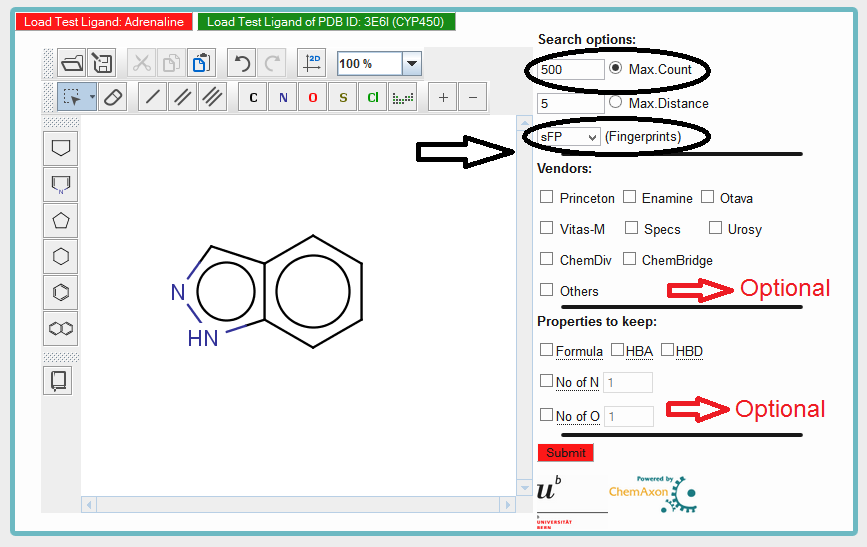
Step 4) Select the Search options
Max Count:It will search for specified number of molecules (as given by count) from database which are most similar to input query molecule (Indazole). In example below the Max Count set to 500.
Max Distance:
It will search for similar molecules from database which are having distance less than or equal to distance specified by user with respect to query molecule(Indazole).
Fingerprints:
Four different fingerprints are available for similarity search calculations. User can choose any one them. For example i have selected "sFP".
Vendors:
It allows you to retrieved the compounds from specific vendors. By default it will retrieved compounds from all vendors. If none of the vendors selected, search will be carried out in default mode. The vendors shown are our own choice and are major contributors to ZINC database.
Properties to keep:
It allows you to specify the properties of molecules which you expected to see in retrieved compounds from database. For example marking "Formula" will only consider the compounds which match molecular formula of query molecule for similarity calculation. "No. of Nitrogen or Oxygen" will make sure that in the retrieved compounds from database you will find specified number of Nitrogen or Oxygen atoms.
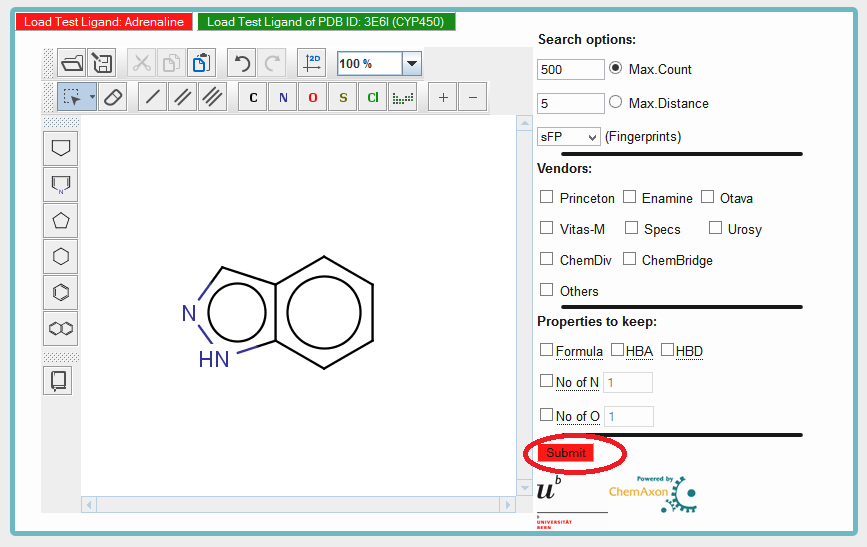
Step 3) Hit the Submit Button and wait for Result
Step 4) Result Window
Result window is shown below. To make it simple to understand the window is divided in to 4 parts which are explained below.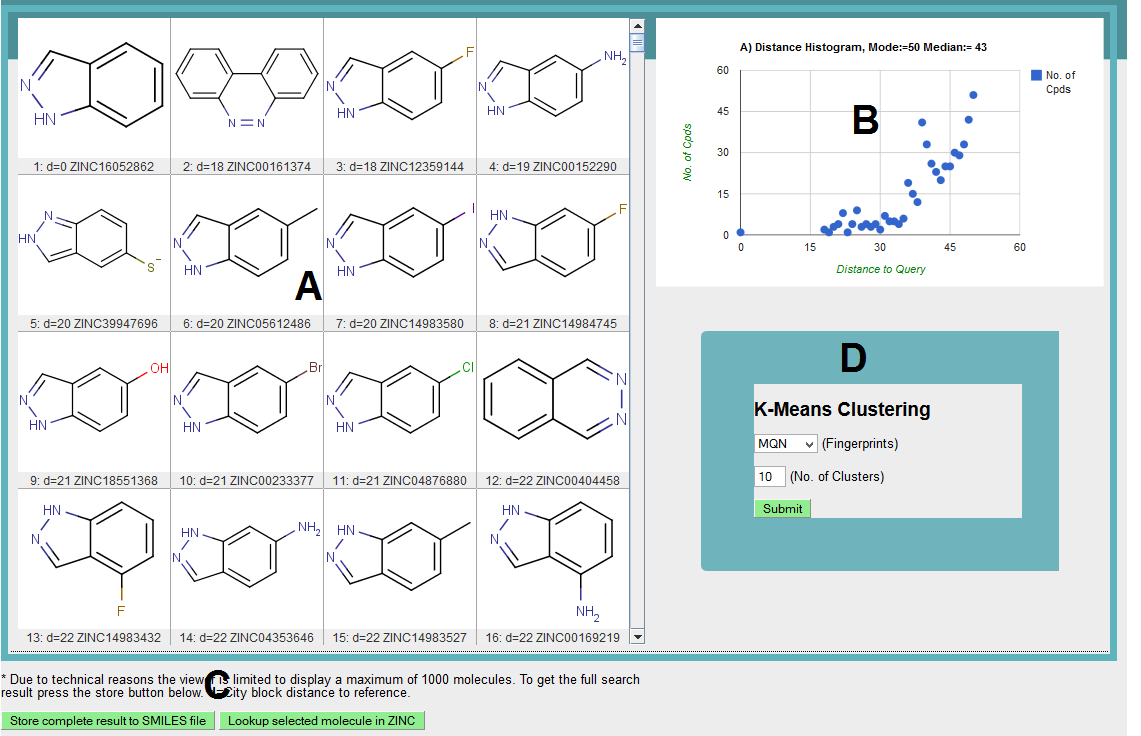
A) Shows the structures of molecules retrieved from the ZINC database using indazole as query molecule. "d" next to each molecule is the distance of molecule to indazole(query).
B) Plot showing distance (panel B, with respect to query) histogram for compounds retrieved from the ZINC database.
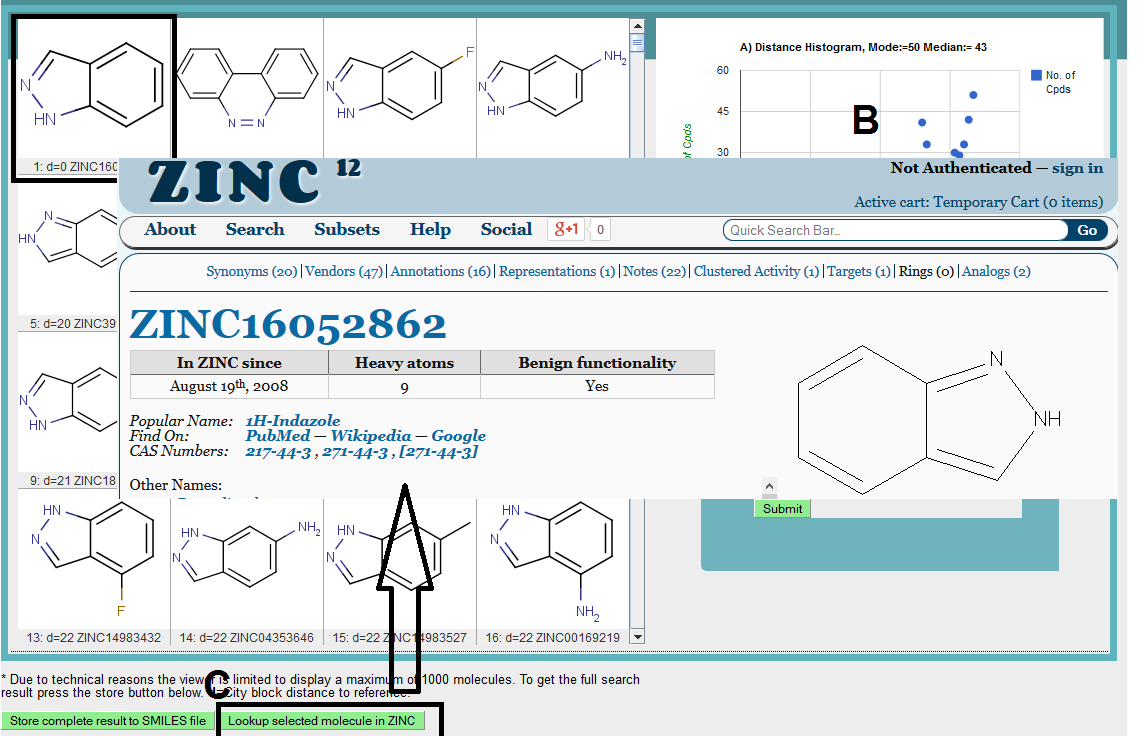
C) Two buttons are available which allows you to store the complete results in smile format and link the displayed molecules (one at time) to parent ZINC database to look for detail information.
D) Option is provided to cluster the nearest neighbors (compounds in window) using K-mean clustering. We need to specify fingerprint (sFP selected) and number of clusters (30). Afterward hit the submit button and wait for results.

Step 5) Display Clusters
Shown below is the 30 clusters obtained for 500 molecules (from above steps) using sFP as fingerprint.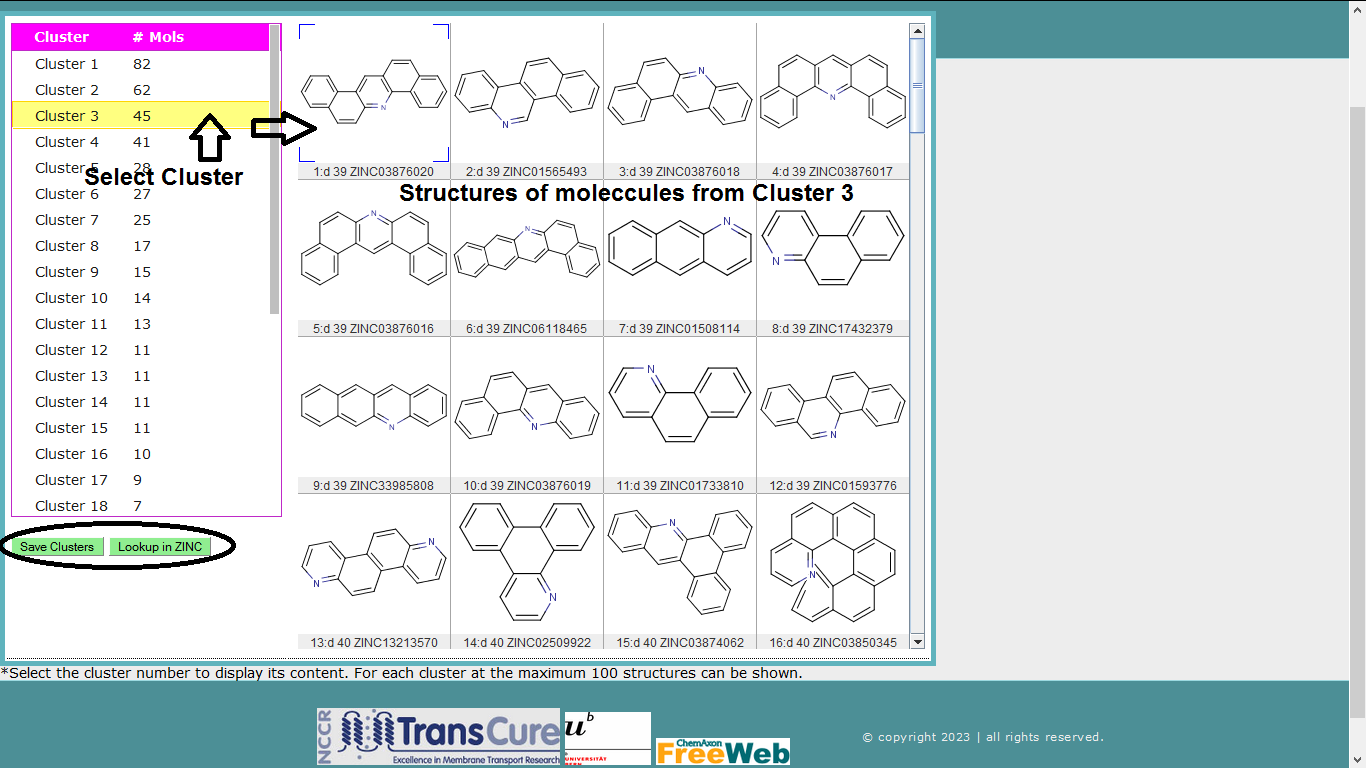
Save Clusters: Molecules save in smile format with annotation of cluster numbers.
Lookup in ZINC: Select molecule and hit this button, it will direct to the ZINC database website and show the information for selected molecule.


