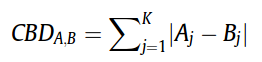-
General Information -
How to Use: Example with Adrenaline -
How to Use: Example with PDB 3E6I -
Workflow -
References -
Problem In Running Application?
General Information
A) Fingerprint
A vector composed of several numerical descriptors of molecular structure and properties.A 1) Substructure Fingerprint (sFP)
The Substructure Fingerprint of a molecule is bit string (a sequence of "0" and "1" digits) that contains information on the structure. Substructure fingerprint (sFP) used in the browser generated using ChemAxon JChem Package with bond length of 7 and contains 1024 bits.The Process of Fingerprint Generation in JChem:
1) Up to a given a bond number (e.g 7), all linear paths (linear patterns) consisting bonds and atoms of a structure are detected.
2) Branching points at the end of each linear pattern are also detected.
3) All cycles (cyclic patterns) are detected.
4) Using a proprietary hashing method, a given number of bits in the bit string are set for each pattern. It is possible, that the same bit is set by multiple patterns. This phenomenon is called bit collision. Few bit collisions in the fingerprint is tolerable, but too many may result in losing information in the fingerprint.
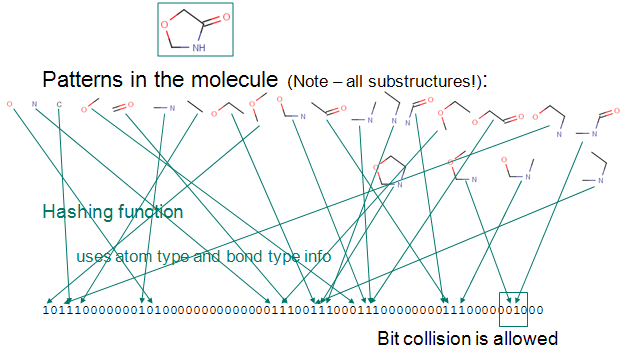
For more details visit, ChemAxon, DAYLIGHT
A 2) Extended-Connectivity Fingerprint (ECFP4)
Extended-Connectivity Fingerprints (ECFPs) are circular topological fingerprints designed for molecular characterization, similarity searching, and structure-activity modeling. They are among the most popular similarity search tools in drug discovery and they are effectively used in a wide variety of applications.Similar to sFP, ECFP4 also encode substructure patterns from molecules on to the bit string of length 1024. Prime Difference in sFP and ECFP4 is the way they percieve the substructure patterns from molecule. Unlike the sFP which considered the linear path upto certain bond lenth, ECFP generate the patterns by considering the atoms into multiple circular layers up to a given diameter. In our browser we have used the ECFP with diameter of 4.
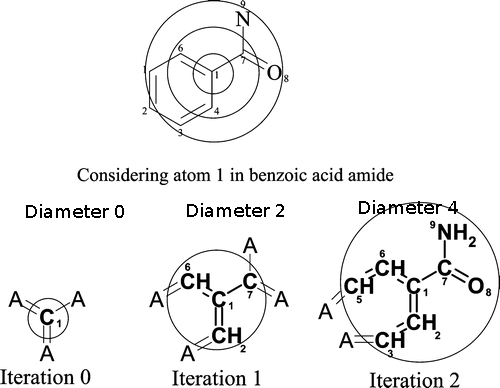
Circular patterns were generated for every atom, followed by hashing of patterns on to the bit string of length 1024. For more details please visit, ChemAxon, ECFP4
A 3) Molecular Quantum Numbers (MQN)
MQN is 42 integer value descriptors of molecular structure, which count atoms, bond types, polar groups, and topological features (shown below).
For more details please visit, MQN
A 3) SMILE Fingerprint (SMIfp)
SMIfp (SMILES fingerprint) is a scalar fingerprint counting the occurances of 34 different symbols in SMILES strings of molecules. The symbols that are counted are summarized below.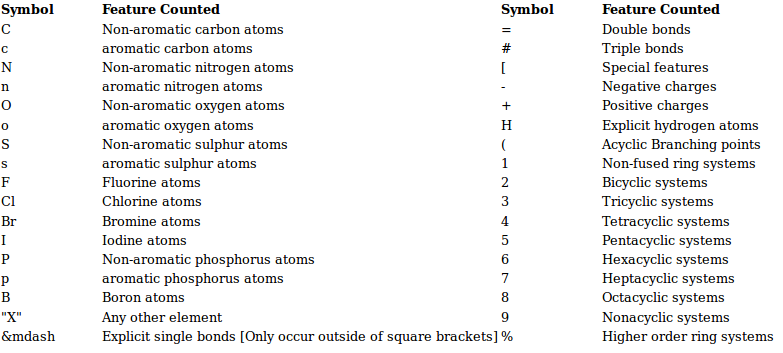
For more details please visit, SMIfp
B) (Dis) Similarity Measure
(Dis) Similarity Measure is a mathematical calculation that quantify similarity or dis-similarity between two objects (molecules). In the browser presented here, we have used City Block Distance (CBD) to calculate dis-similarity (distance) of molecules with respect to given query molecule. The list of retrieved compounds then sorted according to increasing CBD distance with respect to query.Once represented in the form of fingerprint(vector), the City block distance between two molecules(CBD A,B), A and B, with K dimensions is calculated as: